New features of AbiPy v0.7¶
M. Giantomassi and the AbiPy group¶
9th international ABINIT developer workshop
20-22nd May 2019 - Louvain-la-Neuve, Belgium

- These slides have been generated using jupyter, nbconvert and revealjs
- The notebook can be downloaded from this github repo
- To install and configure the software, follow these installation instructions
Use the Space key to navigate through all slides.

What is AbiPy?¶
Python package for:¶
- Generating ABINIT input files automatically
- Post-processing output results (netcdf and text files)
- Interfacing ABINIT with external tools (e.g. Vesta)
- Creating and executing workflows (band structures, DFPT, $GW$, BSE…)
Why python?¶
- Easy to use and to learn
- Great support for science (numpy, scipy, pandas, matplotlib …)
- Interactive environments (ipython, jupyter notebooks, GUIs)
- More powerful and flexible than Fortran for implementing the high-level logic needed in modern ab-initio workflows
- pymatgen ecosystem and the materials project database…
How to install AbiPy¶
Using pip and python wheels:
pip install abipy --user
Using conda (recommended):
conda install abipy --channel abinit
From the github repository (develop mode):
git clone https://github.com/abinit/abipy.git
cd abipy
python setup.py develop
For further info see http://abinit.github.io/abipy/installation.html
AbiPy documentation with galleries of matplotlib examples and workflows¶
In [57]:
%embed https://abinit.github.io/abipy/index.html
Out[57]:
Jupyter notebooks with examples and lessons inspired by the official tutorials¶
In [58]:
%embed https://nbviewer.jupyter.org/github/abinit/abitutorials/blob/master/abitutorials/index.ipynb
Out[58]:

Since AbiPy is not restricted to high-throughput, we'll show how to use the terminal to analyze calculations
Well, a python script would be much more flexible but the goal here is to show that one can replace grep, vim, gnuplot with AbiPy
No perl scripts were harmed in the making of this notebook
Command line interface¶
- abiopen.py ➝ Open output files inside ipython or print/visualize file
- abistruct.py ➝ Operate on crystalline structures read from file
- abicomp.py ➝ Compare multiple files (i.e. convergence studies)
- abiview.py ➝ Quick visualization of output files
- abinp.py ➝ Generate input files for typical calculations
Documentation¶
abistruct.py --helpfor manpageabistruct.py COMMAND --helpfor help aboutCOMMAND
HTML documentation available at http://abinit.github.io/abipy/scripts/index.html
Examples¶
abistruct.py spglib si_scf_GSR.nc
abistruct.py convert si_scf_GSR.nc -f cif
abiopen.py si_scf_GSR.nc --print
and many more...¶
Print results stored in FILE (more than 45 file extensions supported)¶
In [59]:
!abiopen.py si_scf_GSR.nc --print
================================= File Info =================================
Name: si_scf_GSR.nc
Directory: /Users/gmatteo/talks/abipy_intro_abidev2019
Size: 14.83 kb
Access Time: Sun May 19 20:26:25 2019
Modification Time: Sun May 12 04:20:35 2019
Change Time: Sun May 12 04:20:35 2019
================================= Structure =================================
Full Formula (Si2)
Reduced Formula: Si
abc : 3.866975 3.866975 3.866975
angles: 60.000000 60.000000 60.000000
Sites (2)
# SP a b c cartesian_forces
--- ---- ---- ---- ---- -----------------------------------------------------------
0 Si 0 0 0 [-5.89948302e-27 -1.93366148e-27 2.91016902e-27] eV ang^-1
1 Si 0.25 0.25 0.25 [ 5.89948302e-27 1.93366148e-27 -2.91016902e-27] eV ang^-1
Abinit Spacegroup: spgid: 227, num_spatial_symmetries: 48, has_timerev: True, symmorphic: True
Stress tensor (Cartesian coordinates in GPa):
[7.86452261e-11 5.21161758e+00 0.00000000e+00]
[0.00000000e+00 0.00000000e+00 5.21161758e+00]]
Pressure: -5.212 (GPa)
Energy: -241.23646832 (eV)
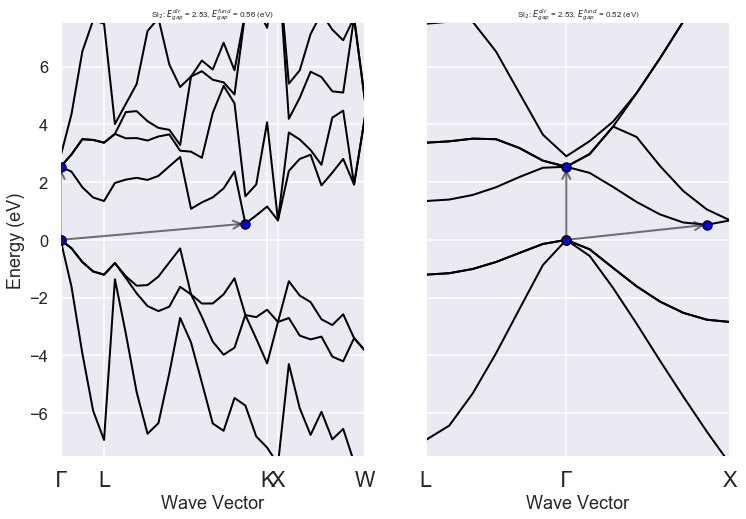
============================== Electronic Bands ==============================
Number of electrons: 8.0, Fermi level: 5.598 (eV)
nsppol: 1, nkpt: 29, mband: 8, nspinor: 1, nspden: 1
smearing scheme: none, tsmear_eV: 0.272, occopt: 1
Direct gap:
Energy: 2.532 (eV)
Initial state: spin=0, kpt=[+0.000, +0.000, +0.000], weight: 0.002, band=3, eig=5.598, occ=2.000
Final state: spin=0, kpt=[+0.000, +0.000, +0.000], weight: 0.002, band=4, eig=8.130, occ=0.000
Fundamental gap:
Energy: 0.562 (eV)
Initial state: spin=0, kpt=[+0.000, +0.000, +0.000], weight: 0.002, band=3, eig=5.598, occ=2.000
Final state: spin=0, kpt=[+0.375, +0.375, +0.000], weight: 0.012, band=4, eig=6.161, occ=0.000
Bandwidth: 11.856 (eV)
Valence maximum located at:
spin=0, kpt=[+0.000, +0.000, +0.000], weight: 0.002, band=3, eig=5.598, occ=2.000
Conduction minimum located at:
spin=0, kpt=[+0.375, +0.375, +0.000], weight: 0.012, band=4, eig=6.161, occ=0.000
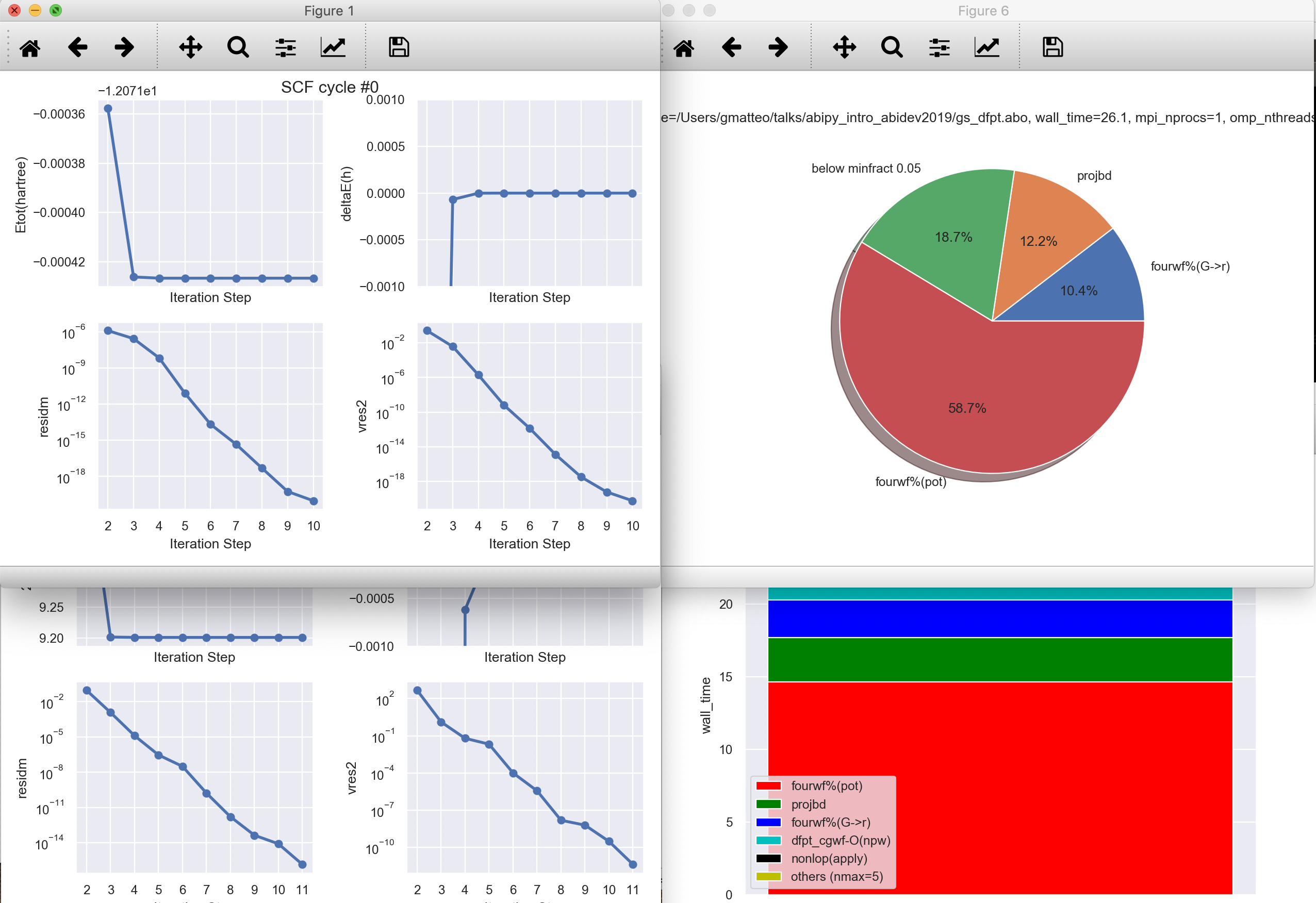
To produce a predefined set of matplotlib figures, use:¶
abiopen.py mgb2_kpath_FATBANDS.nc --expose --seaborn
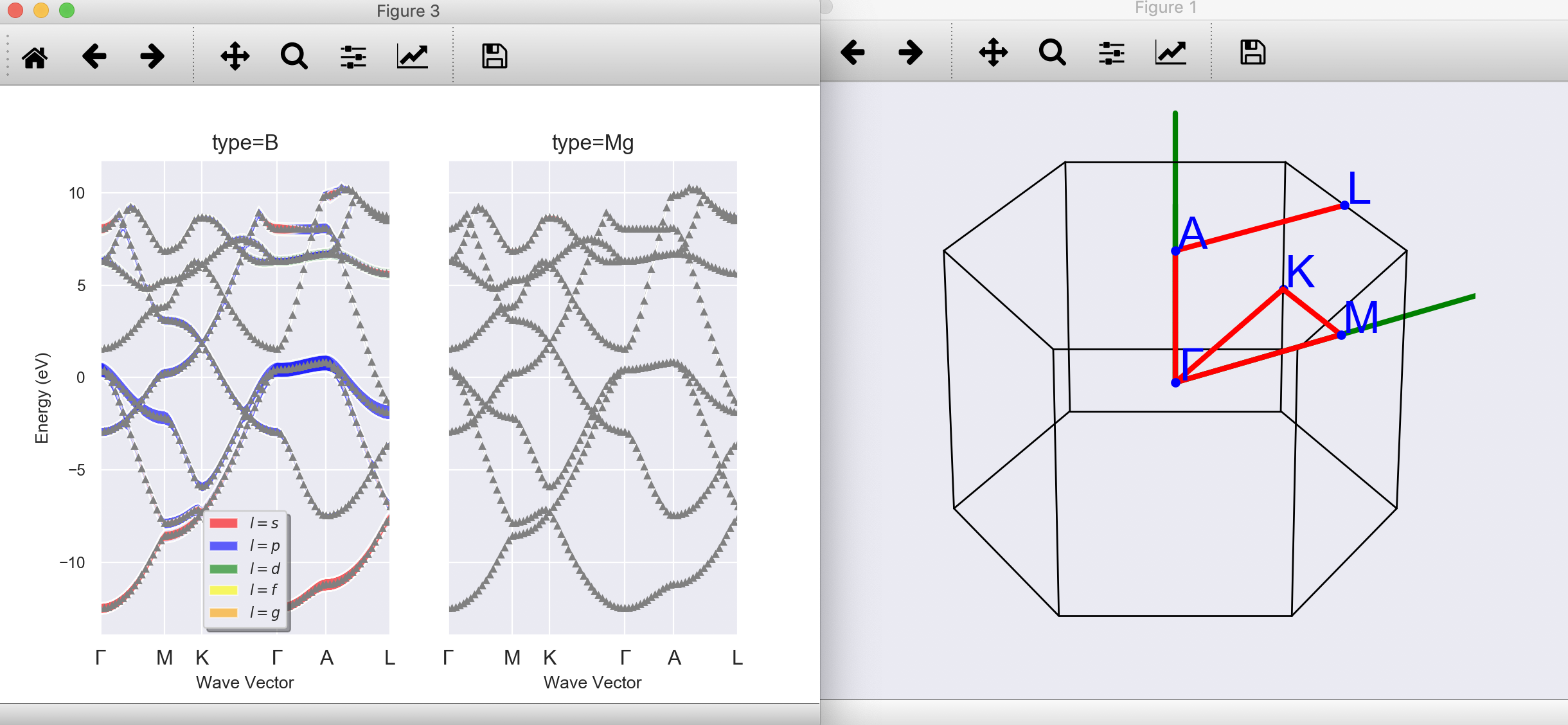
Replace --expose with --notebook to generate a jupyter notebook with predefined python code¶
abiopen.py supports output files (note abo extension):¶
In [60]:
!abiopen.py gs_dfpt.abo -p
ndtset: 3, completed: True
Full Formula (C2)
Reduced Formula: C
abc : 2.508336 2.508336 2.508336
angles: 60.000000 60.000000 60.000000
Sites (2)
# SP a b c
--- ---- ---- ---- ----
0 C 0 0 0
1 C 0.25 0.25 0.25
Abinit Spacegroup: spgid: 227, num_spatial_symmetries: 48, has_timerev: True, symmorphic: True
========================= Dimensions of calculation =========================
intxc ionmov iscf lmnmax lnmax mgfft mpssoang mqgrid natom \
dataset
1 0 0.0 7 1 1 24 2 3001 2
2 0 NaN 7 1 1 24 2 3001 2
3 0 NaN 7 1 1 24 2 3001 2
nloc_mem nspden nspinor nsppol nsym n1xccc ntypat occopt \
dataset
1 1 1 1 1 48 2501 1 1
2 1 1 1 1 48 2501 1 1
3 1 1 1 1 48 2501 1 1
xclevel mband mffmem mkmem mpw nfft nkpt mem_per_proc_mb \
dataset
1 1 4 1 8 609 13824 8 4.791
2 1 4 1 64 609 13824 64 11.529
3 1 4 1 64 609 13824 64 12.285
wfk_size_mb denpot_size_mb spg_symbol spg_number \
dataset
1 0.299 0.107 Fd-3m 227
2 2.381 0.107 Fd-3m 227
3 2.381 0.107 Fd-3m 227
bravais
dataset
1 Bravais cF (face-center cubic)
2 Bravais cF (face-center cubic)
3 Bravais cF (face-center cubic)
and log files as well:¶
In [61]:
!abiopen.py run.log -p
Events found in /Users/gmatteo/talks/abipy_intro_abidev2019/run.log [1] <AbinitComment at m_dtfil.F90:1082> Output file: run.abo already exists. [2] <AbinitComment at m_dtfil.F90:1106> Renaming old run.abo to run.abo0001 [3] <AbinitWarning at m_mpinfo.F90:2295> nkpt*nsppol (29) is not a multiple of nproc_kpt (2) The k-point parallelisation is not efficient. [4] <AbinitWarning at m_mpinfo.F90:2295> nkpt*nsppol (29) is not a multiple of nproc_kpt (2) The k-point parallelisation is not efficient. [5] <AbinitComment at m_xgScalapack.F90:237> xgScalapack in auto mode [6] <AbinitWarning at m_phonons.F90:3215> ph_nqpath <= 0 or ph_ndivsm <= 0. Phonon bands won't be produced. returning [7] <AbinitComment at m_xgScalapack.F90:237> xgScalapack in auto mode [8] <AbinitWarning at m_phonons.F90:3215> ph_nqpath <= 0 or ph_ndivsm <= 0. Phonon bands won't be produced. returning num_errors: 0, num_warnings: 4, num_comments: 4, completed: True

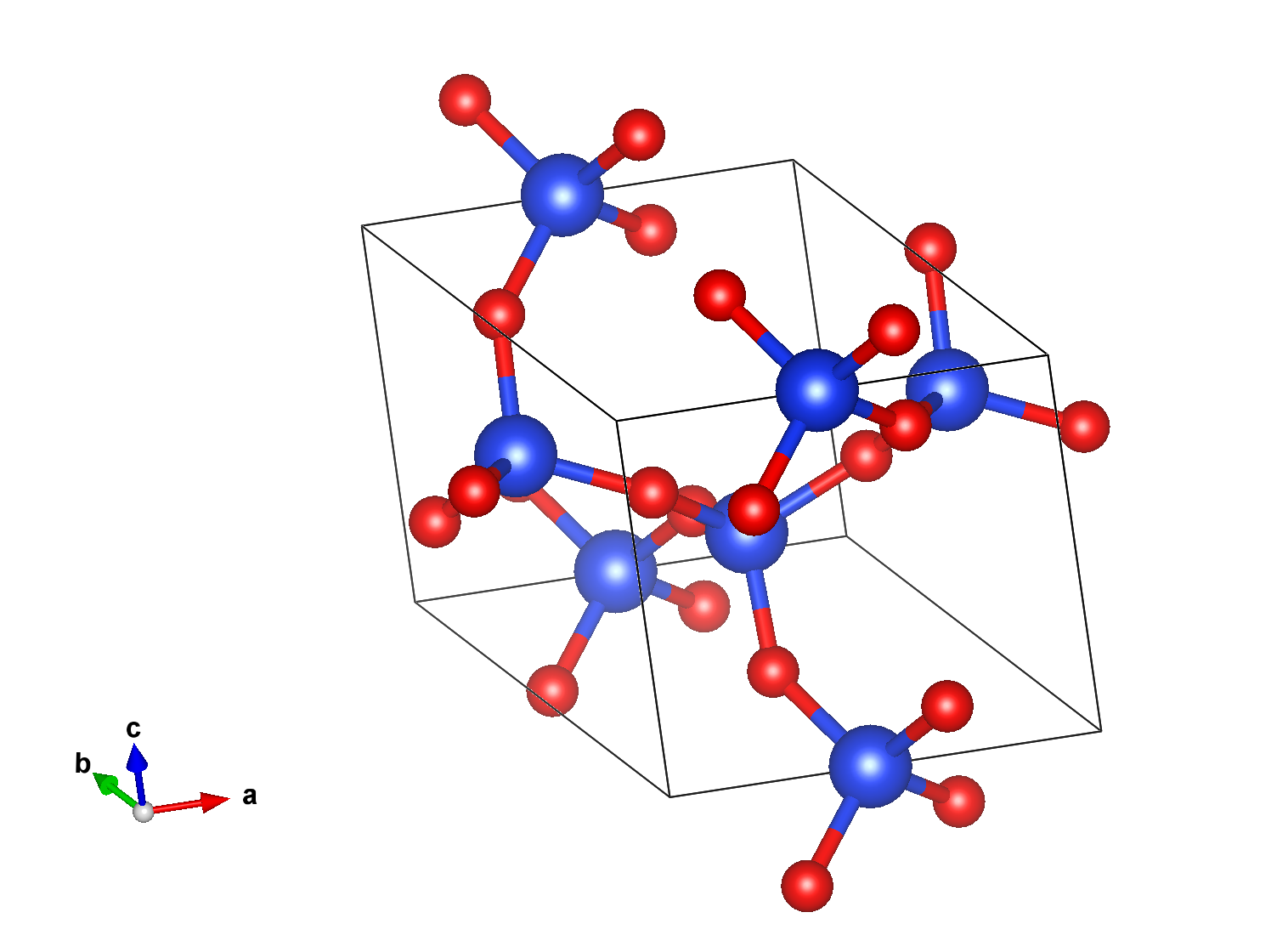
It works with any file providing a structure object (.nc, .abi, .cif …)¶
Convert structure from netcdf format to CIF (abivars, xsf, poscar, qe, siesta, wannier90, json)¶
In [62]:
!abistruct.py convert si_scf_GSR.nc -f cif
# generated using pymatgen data_Si _symmetry_space_group_name_H-M 'P 1' _cell_length_a 3.86697464 _cell_length_b 3.86697464 _cell_length_c 3.86697464 _cell_angle_alpha 60.00000000 _cell_angle_beta 60.00000000 _cell_angle_gamma 60.00000000 _symmetry_Int_Tables_number 1 _chemical_formula_structural Si _chemical_formula_sum Si2 _cell_volume 40.88829228 _cell_formula_units_Z 2 loop_ _symmetry_equiv_pos_site_id _symmetry_equiv_pos_as_xyz 1 'x, y, z' loop_ _atom_site_type_symbol _atom_site_label _atom_site_symmetry_multiplicity _atom_site_fract_x _atom_site_fract_y _atom_site_fract_z _atom_site_occupancy Si Si1 1 0.000000 0.000000 0.000000 1 Si Si2 1 0.250000 0.250000 0.250000 1
Connect to the materials project database to find similar structures.¶
In [66]:
!abistruct.py mp_match si_scf_GSR.nc
# Found 1 structures in Materials Project database (use `verbose` to get further info)
########################## abivars input for mp-149 ##########################
# Full Formula (Si2)
# Reduced Formula: Si
# abc : 3.866975 3.866975 3.866975
# angles: 60.000000 60.000000 60.000000
#
# Spglib space group info (magnetic symmetries not taken into account).
# Spacegroup: Fd-3m (227), Hall: F 4d 2 3 -1d, Abinit spg_number: None
# Crystal_system: cubic, Lattice_type: cubic, Point_group: m-3m
#
# Idx Symbol Reduced_Coords Wyckoff EqIdx
# ----- -------- -------------------------- --------- -------
# 0 Si +0.87500 +0.87500 +0.87500 (2a) 0
# 1 Si +0.12500 +0.12500 +0.12500 (2a) 0
natom 2
ntypat 1
typat 1 1
znucl 14
xred
0.8750000000 0.8750000000 0.8750000000
0.1250000000 0.1250000000 0.1250000000
acell 1.0 1.0 1.0
rprim
6.3285005334 0.0000000000 3.6537614829
2.1095001778 5.9665675247 3.6537614829
0.0000000000 0.0000000000 7.3075229659
To search by chemical system or formula (no FILE required)¶
In [67]:
!abistruct.py mp_search LiF
# Found 3 structures in Materials Project database (use `verbose` to get further info)
pretty_formula e_above_hull energy_per_atom \
mp-1185301 LiF 0.007565 -4.837577
mp-1009009 LiF 0.273111 -4.572031
mp-1138 LiF 0.000000 -4.845142
formation_energy_per_atom nsites volume spacegroup.symbol \
mp-1185301 -3.173315 4 42.223600 P6_3mc
mp-1009009 -2.907769 2 16.768040 Pm-3m
mp-1138 -3.180880 2 17.022154 Fm-3m
spacegroup.number band_gap total_magnetization material_id \
mp-1185301 186 7.4778 0.000000e+00 mp-1185301
mp-1009009 221 7.5593 0.000000e+00 mp-1009009
mp-1138 225 8.7161 9.000000e-07 mp-1138
a b c alpha beta gamma
mp-1185301 3.139544 3.139544 4.946425 90.0 90.0 120.000008
mp-1009009 2.559533 2.559533 2.559533 90.0 90.0 90.000000
mp-1138 2.887419 2.887419 2.887419 60.0 60.0 60.000000
######################## abivars input for mp-1185301 ########################
# Full Formula (Li2 F2)
# Reduced Formula: LiF
# abc : 3.139544 3.139544 4.946425
# angles: 90.000000 90.000000 120.000008
#
# Spglib space group info (magnetic symmetries not taken into account).
# Spacegroup: P6_3mc (186), Hall: P 6c -2c, Abinit spg_number: None
# Crystal_system: hexagonal, Lattice_type: hexagonal, Point_group: 6mm
#
# Idx Symbol Reduced_Coords Wyckoff EqIdx
# ----- -------- -------------------------- --------- -------
# 0 Li +0.00000 +0.00000 +0.38019 (2b) 0
# 1 Li +0.33333 +0.66667 +0.88019 (2b) 0
# 2 F +0.00000 +0.00000 +0.99481 (2b) 2
# 3 F +0.33333 +0.66667 +0.49481 (2b) 2
natom 4
ntypat 2
typat
1 1 2
2
znucl 3 9
xred
0.0000000000 0.0000000000 0.3801910000
0.3333333333 0.6666666667 0.8801900000
0.0000000000 0.0000000000 0.9948070000
0.3333333333 0.6666666667 0.4948100000
acell 1.0 1.0 1.0
rprim
5.9328791503 0.0000000000 0.0000000000
-2.9664403365 5.1380236222 0.0000000000
0.0000000000 0.0000000000 9.3473877186
######################## abivars input for mp-1009009 ########################
# Full Formula (Li1 F1)
# Reduced Formula: LiF
# abc : 2.559533 2.559533 2.559533
# angles: 90.000000 90.000000 90.000000
#
# Spglib space group info (magnetic symmetries not taken into account).
# Spacegroup: Pm-3m (221), Hall: -P 4 2 3, Abinit spg_number: None
# Crystal_system: cubic, Lattice_type: cubic, Point_group: m-3m
#
# Idx Symbol Reduced_Coords Wyckoff EqIdx
# ----- -------- -------------------------- --------- -------
# 0 Li +0.00000 +0.00000 +0.00000 (1a) 0
# 1 F +0.50000 +0.50000 +0.50000 (1b) 1
natom 2
ntypat 2
typat 1 2
znucl 3 9
xred
0.0000000000 0.0000000000 0.0000000000
0.5000000000 0.5000000000 0.5000000000
acell 1.0 1.0 1.0
rprim
4.8368167948 0.0000000000 0.0000000000
0.0000000000 4.8368167948 0.0000000000
0.0000000000 0.0000000000 4.8368167948
######################### abivars input for mp-1138 #########################
# Full Formula (Li1 F1)
# Reduced Formula: LiF
# abc : 2.887419 2.887419 2.887419
# angles: 60.000000 60.000000 60.000000
#
# Spglib space group info (magnetic symmetries not taken into account).
# Spacegroup: Fm-3m (225), Hall: -F 4 2 3, Abinit spg_number: None
# Crystal_system: cubic, Lattice_type: cubic, Point_group: m-3m
#
# Idx Symbol Reduced_Coords Wyckoff EqIdx
# ----- -------- -------------------------- --------- -------
# 0 Li +0.00000 +0.00000 +0.00000 (1a) 0
# 1 F +0.50000 +0.50000 +0.50000 (1b) 1
natom 2
ntypat 2
typat 1 2
znucl 3 9
xred
0.0000000000 0.0000000000 0.0000000000
0.5000000000 0.5000000000 0.5000000000
acell 1.0 1.0 1.0
rprim
4.7254082787 0.0000000000 2.7282157318
1.5751360851 4.4551576322 2.7282157281
0.0000000000 0.0000000000 5.4564314785
Need to call anaddb to compute and visualize ph-bands and DOS from DDB?¶
abiview.py ddb ZnSe_hex_qpt_DDB --seaborn
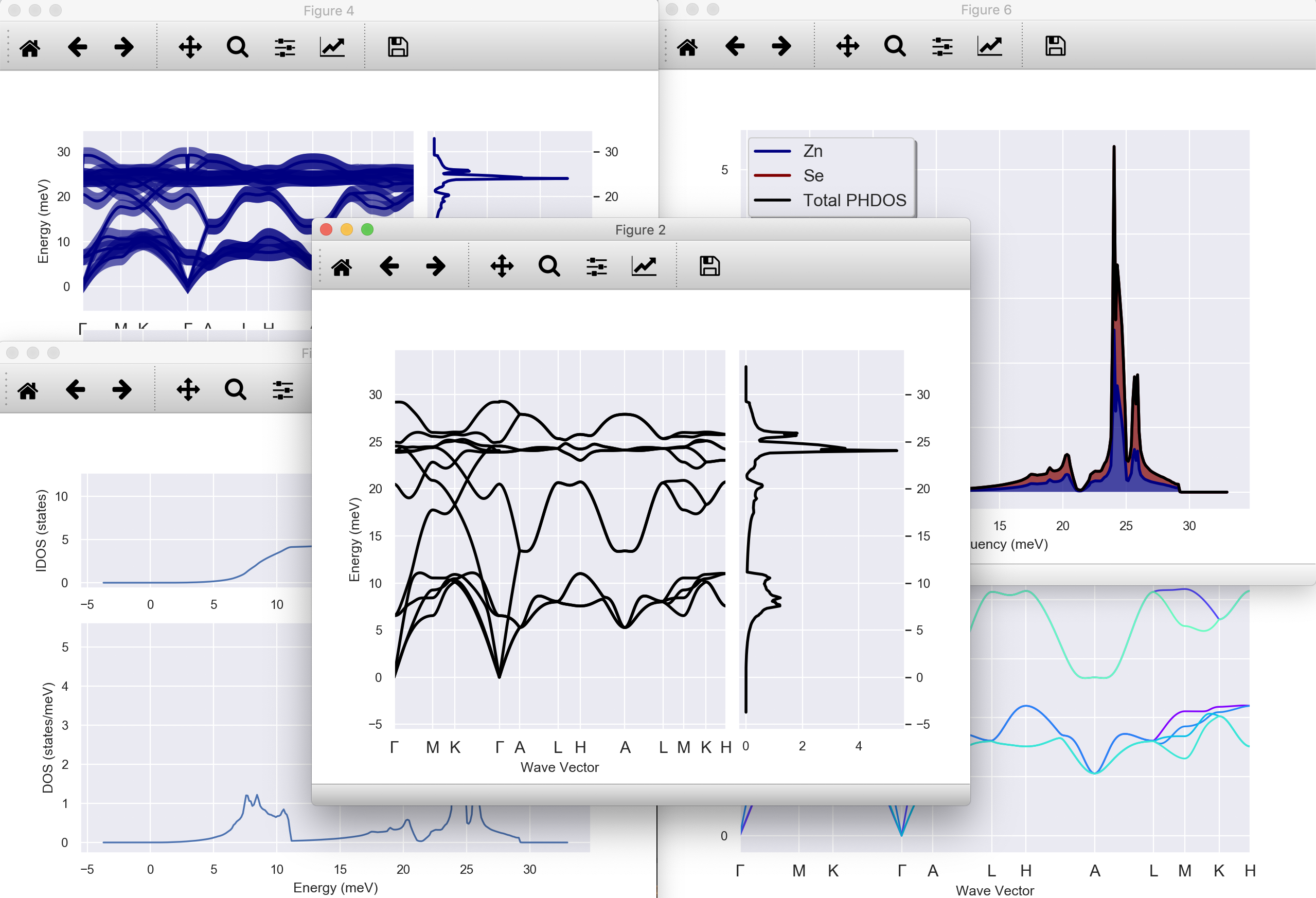
Add --phononwebsite to visualize data on the phononwebsite by Henrique¶

To compare multiple structures:¶
In [68]:
!abicomp.py structure *.cif si_nscf_GSR.nc `find . -name *_DDB`
Spglib options: symprec= 0.001 angle_tolerance= 5.0
Lattice parameters:
formula natom alpha beta gamma a b \
AlAs.cif Al1 As1 2 60.0 60.0 60.0 3.970101 3.970101
si.cif Si2 2 60.0 60.0 60.0 3.866975 3.866975
si_nscf_GSR.nc Si2 2 60.0 60.0 60.0 3.866975 3.866975
ZnSe_hex_qpt_DDB Zn2 Se2 4 90.0 90.0 120.0 4.050187 4.050187
c volume abispg_num spglib_symb spglib_num \
AlAs.cif 3.970101 44.247584 None F-43m 216
si.cif 3.866975 40.888292 None Fd-3m 227
si_nscf_GSR.nc 3.866975 40.888292 227 Fd-3m 227
ZnSe_hex_qpt_DDB 6.652328 94.504936 0 P6_3mc 186
spglib_lattice_type
AlAs.cif cubic
si.cif cubic
si_nscf_GSR.nc cubic
ZnSe_hex_qpt_DDB hexagonal
Note shell wildcard characters and Unix find inside backticks (bash rocks!)¶
In [70]:
paths = [
"mgb2_888k_0.01tsmear_DDB",
"mgb2_888k_0.04tsmear_DDB",
"mgb2_121212k_0.01tsmear_DDB",
"mgb2_121212k_0.04tsmear_DDB",
]
paths = [os.path.join(abidata.dirpath, "refs", "mgb2_phonons_nkpt_tsmear", f)
for f in paths]
robot = abilab.DdbRobot()
for i, path in enumerate(paths):
robot.add_file(path, path)
In [71]:
# Define function to change labels:
func = lambda ddb: "nkpt: %s, tsmear: %.2f" % (
ddb.header["nkpt"], ddb.header["tsmear"])
robot.remap_labels(func)
robot
Out[71]:
- nkpt: 256, tsmear: 0.01
- nkpt: 256, tsmear: 0.04
- nkpt: 864, tsmear: 0.01
- nkpt: 864, tsmear: 0.04
Now we can build a dataframe with the most important parameters:¶
In [72]:
robot.get_params_dataframe()
Out[72]:
| nkpt | nsppol | ecut | tsmear | occopt | ixc | nband | usepaw | |
|---|---|---|---|---|---|---|---|---|
| nkpt: 256, tsmear: 0.01 | 256 | 1 | 35.0 | 0.01 | 4 | 1 | 8 | 0 |
| nkpt: 256, tsmear: 0.04 | 256 | 1 | 35.0 | 0.04 | 4 | 1 | 8 | 0 |
| nkpt: 864, tsmear: 0.01 | 864 | 1 | 35.0 | 0.01 | 4 | 1 | 8 | 0 |
| nkpt: 864, tsmear: 0.04 | 864 | 1 | 35.0 | 0.04 | 4 | 1 | 8 | 0 |
and check that all DDBs have been computed with the same crystalline structure:¶
In [73]:
robot.get_lattice_dataframe()
Out[73]:
| formula | natom | alpha | beta | gamma | a | b | c | volume | abispg_num | spglib_symb | spglib_num | spglib_lattice_type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nkpt: 256, tsmear: 0.01 | Mg1 B2 | 3 | 90.0 | 90.0 | 120.0 | 3.086 | 3.086 | 3.523 | 29.055953 | 0 | P6/mmm | 191 | hexagonal |
| nkpt: 256, tsmear: 0.04 | Mg1 B2 | 3 | 90.0 | 90.0 | 120.0 | 3.086 | 3.086 | 3.523 | 29.055953 | 0 | P6/mmm | 191 | hexagonal |
| nkpt: 864, tsmear: 0.01 | Mg1 B2 | 3 | 90.0 | 90.0 | 120.0 | 3.086 | 3.086 | 3.523 | 29.055953 | 0 | P6/mmm | 191 | hexagonal |
| nkpt: 864, tsmear: 0.04 | Mg1 B2 | 3 | 90.0 | 90.0 | 120.0 | 3.086 | 3.086 | 3.523 | 29.055953 | 0 | P6/mmm | 191 | hexagonal |
To analyze the effect of k-sampling/smearing on the vibrational properties:¶
In [74]:
# Invoke anaddb and store results
r = robot.anaget_phonon_plotters(nqsmall=2)
r.phbands_plotter.gridplot_with_hue("tsmear", with_dos=True);

Input file for band structure calculation + DOS¶
- GS run to get the density
- NSCF run along high-symmetry k-path
- NSCF run with k-mesh to compute the DOS
In [77]:
multi = abilab.ebands_input(structure="si.cif",
pseudos="14si.pspnc",
ecut=8,
spin_mode="unpolarized",
smearing=None,
dos_kppa=5000)
multi.get_vars_dataframe("kptopt", "iscf", "ngkpt")
Out[77]:
| kptopt | iscf | ngkpt | |
|---|---|---|---|
| dataset 0 | 1 | None | [8, 8, 8] |
| dataset 1 | -11 | -2 | None |
| dataset 2 | 1 | -2 | [14, 14, 14] |
To build an input for SCF+NSCF run with (relaxed) structure from the materials project database:¶
abinp.py ebands mp-149 abinp.py is handy for generating input files quickly but it cannot compete with the flexibility of the python interface:¶
In [79]:
def make_scf_input(ecut=2, ngkpt=(4, 4, 4)):
"""
Generate an `AbinitInput` to perform GS calculation for AlAs.
Args:
ecut: Cutoff energy in Ha.
ngkpt: k-mesh divisions
Return:
`AbinitInput` object
"""
gs_inp = abilab.AbinitInput(structure="AlAs.cif",
pseudos=["13al.pspnc", "33as.pspnc"])
# Set the value of the Abinit variables needed for GS runs.
gs_inp.set_vars(
nband=4,
ecut=ecut,
ngkpt=ngkpt,
nshiftk=4,
shiftk=[0.0, 0.0, 0.5, # This gives the usual fcc Monkhorst-Pack grid
0.0, 0.5, 0.0,
0.5, 0.0, 0.0,
0.5, 0.5, 0.5],
tolvrs=1.0e-10,
)
return gs_inp
Inside the notebook, one gets the HTML representation with links to the documentation:¶
In [80]:
make_scf_input()
Out[80]:
############################################################################################
# SECTION: basic
############################################################################################
nband 4
ecut 2
ngkpt 4 4 4
nshiftk 4
shiftk
0.0 0.0 0.5
0.0 0.5 0.0
0.5 0.0 0.0
0.5 0.5 0.5
tolvrs 1e-10
############################################################################################
# STRUCTURE
############################################################################################
natom 2
ntypat 2
typat 1 2
znucl 13 33
xred
0.0000000000 0.0000000000 0.0000000000
0.2500000000 0.2500000000 0.2500000000
acell 1.0 1.0 1.0
rprim
6.4972715500 0.0000000000 3.7512014784
2.1657571833 6.1256863630 3.7512014784
0.0000000000 0.0000000000 7.5024029568
# SECTION: basic
############################################################################################
nband 4
ecut 2
ngkpt 4 4 4
nshiftk 4
shiftk
0.0 0.0 0.5
0.0 0.5 0.0
0.5 0.0 0.0
0.5 0.5 0.5
tolvrs 1e-10
############################################################################################
# STRUCTURE
############################################################################################
natom 2
ntypat 2
typat 1 2
znucl 13 33
xred
0.0000000000 0.0000000000 0.0000000000
0.2500000000 0.2500000000 0.2500000000
acell 1.0 1.0 1.0
rprim
6.4972715500 0.0000000000 3.7512014784
2.1657571833 6.1256863630 3.7512014784
0.0000000000 0.0000000000 7.5024029568
Moreover, from an AbiniInput one can easily build more complicate workflows:¶
In [81]:
def build_flow_alas_phonons():
"""
Build and return a Flow to compute the dynamical matrix on a (2, 2, 2) qmesh
as well as DDK and Born effective charges.
The final DDB with all perturbations will be merged automatically and placed
in the Flow `outdir` directory.
"""
from abipy import flowtk
scf_input = make_scf_input(ecut=6, ngkpt=(4, 4, 4))
return flowtk.PhononFlow.from_scf_input("flow_alas_phonons", scf_input,
ph_ngqpt=(2, 2, 2), with_becs=True)
Abipy will call Abinit to get the list of DFPT perturbations and…¶
In [82]:
flow_phbands = build_flow_alas_phonons()
flow_phbands.get_graphviz()
Out[82]:
Future developments¶
Post-processing tools¶
- Support for more netcdf files
- More post-processing tools for MD calculations
- More converters and interfaces for third-party applications
- Integrate AbiPy with jupyterlab to create a flexible graphical enviroment for Abinit exposing (part) of the python API
- Explore web-based technologies for data analysis and visualization (plotly, dash)
- Develop toolkit to build web apps powered by AbiPy and pymatgen to disseminate scientific results.
Continuous Integration¶
Use AbiPy programmatic interface to implement:
- Validation of parallel algorithms for np in range(1, N)
- Stress testing
- Benchmarks